Highlighted papers
For a complete list of papers, take a look at Gautam’s Google Scholar or ORCID profiles.
We read, review and post preprints.
Lab Google scholar profiles: Jana Helsen | Hiral Shah
At EMBL
Maslennikova et al.
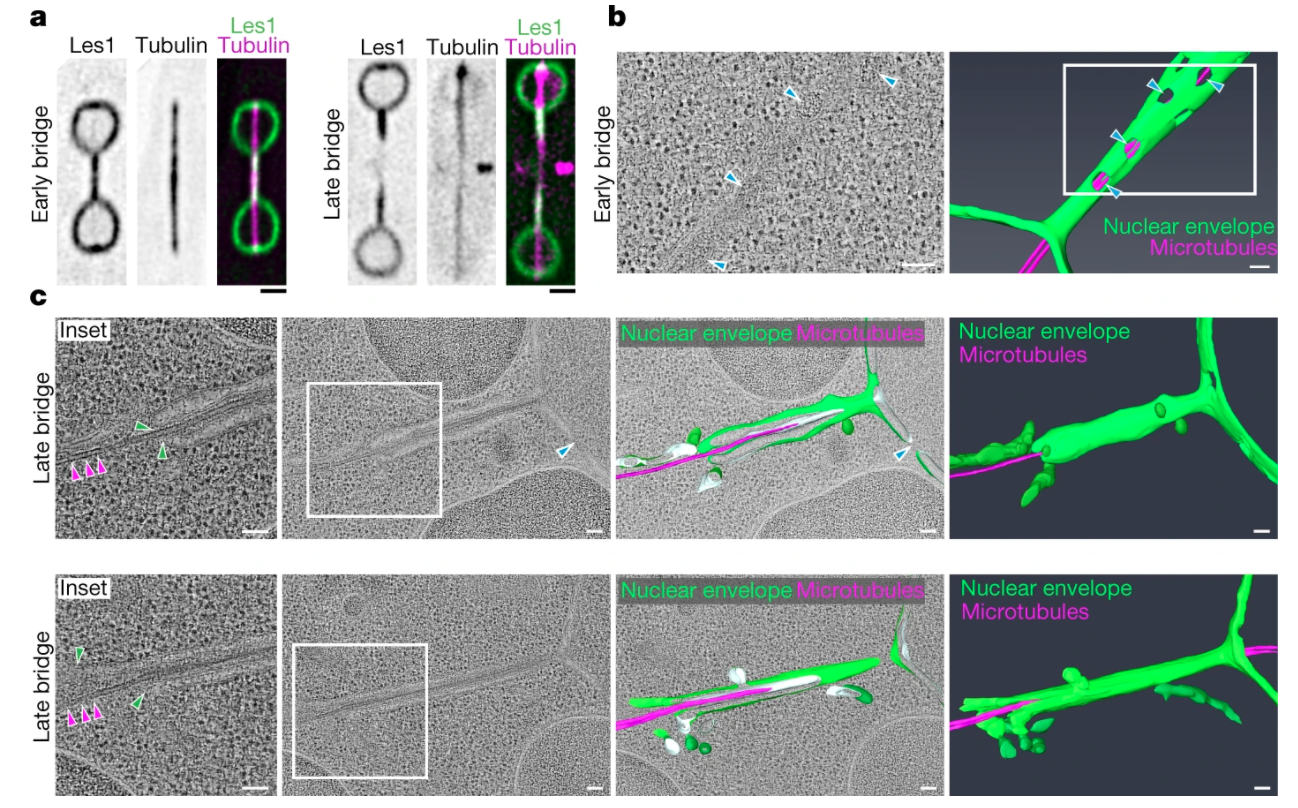
Dystonia-associated Torsins sustain CLCC1 function to promote membrane fusion of the nuclear envelope for NPC biogenesis
bioRxiv, 2025 DOI: 10.1101/2025.11.07.687155
DYT1 early-onset dystonia is a severe, incurable disorder of the central nervous system caused by mutations in the gene encoding Torsin1A (Tor1A, DYT1), an ER-resident ATPase with roles in lipid metabolism and NPC biogenesis. However, their function underlying the disease pathology along with their phylogenetic distribution are not understood
In this study, we show that fly germ cells without Torsins are arrested in their development. We use proximity labelling in Human cells to show that CLCC1 is a novel binding partner for Torsin. Overexpression of CLCC1 in both humans and flies is sufficient to fix problems in NPC biogenesis rising from Torsin deletions. We use Hidden Markov Models to map the phylogeny of Torsins and CLCC1 and show that they are universally present in Holozoa.
This study is a collaboration with the Labs of Stefano Vanni, Madhav Jagannathan, Ulrike Kutay,
Fischer, Wojtynek, Kumar et al.
A conserved mechanism of membrane fusion in nuclear pore complex assembly
In eukaryotes, all transport between the cytoplasm and the nucleus happens through the nuclear pore complex (NPC), a ubiquitous, conserved structure that is present across both the inner and outer nuclear membranes. When NPC biogenesis occurs, eukaryotes need a mechanism to insert the NPC into the nuclear membrane. To successfully insert the NPC into the nuclear membrane, the inner and outer membranes need to undergo fusion. This fusion step occurs in all eukaryotes, and we do not know the molecular mechanism behind this.
In this study, we show that two membrane proteins, Brl1 and Brr6, mediate this mechanism in S. cerevisiae. Using molecular dynamics simulations, we show that these two proteins drive membrane fusion by forming a channel across bilayers that enables lipid exchange. We use phylogenetic analyses to show that these two proteins are broadly conserved across eukaryotes. We also show that CLCC1 in H. sapiens and D. melanogaster is the functional equivalent of these two proteins that works through a similar mechanism.
This preprint is a result from the collaboration between us and the labs of Madhav Jagannathan, Ulrike Kutay, Stefano Vanni, and Karsten Weis.
Diatom ultrastructural diversity across controlled and natural environments
Current Biology, 2025 DOI:10.1016/j.cub.2025.10.024
bioRxiv, 2025 DOI: 10.1101/2025.06.16.659906
Diatoms are microalgae which play critical roles in our planet’s environment. Despite their importance, imaging diatoms has proved to be very difficult due to their robust silica cell wall that blocks the entry of most dyes and gene editing tools.
In this study, we demonstrate that cryo-fixation combined with ultrastructural expansion microscopy (cryo-ExM) can overcome the silica barrier across diverse diatoms spanning over 80 million years of evolutionary time. We derive insights about the microtubule organization, photosynthetic diversity, and diatom symbioses.
This study is a collaboration with the Vincent lab, Schwab lab, Dudin lab, Yeh lab, and the EMBL Mobile Labs
Elliott et al.
Repulsive particle interactions enable selective information processing at cellular interfaces
Physical Review Letters, 2025 DOI: 10.1103/ywr6-mzz5
aRxiv, 2025 DOI: 10.1101/2024.02.20.581313
Living systems constantly adapt to changing environmental conditions. They achieve this by processing information. Several principles governing information processing in living systems have been identified in gene regulation, intra and intercellular signaling, and cell patterning. Physical interactions, mechanical properties, and geometric relations also provide inputs for information processing. However, the principles behind such physically-mediated information processing are not understood.
Here, we investigate a type of physically-mediated information processing: particles embedded in interfaces of membrane-enclosed systems. We demonstrate that particle repulsion results in the selective transmission of information across the membrane, and compare our theory to a living system: the distribution of nuclear pore complexes embedded in the nuclear membrane of Spaheroforma arctica and their interactions with the local cytoskeleton.
This preprint is a result from the collaboration between us and the lab of Anna Erzberger.
Helsen J et al.
Centromeres evolve progressively through selection at the kinetochore interface
Nature (accepted)
bioRxiv, 2025 DOI: 10.1101/2025.01.16.633479
This work investigates the fundamental evolutionary principles underlying transitions in the point centromeres of the Saccharomycetaceae spp.
We developed PCAn to systematically map point centromeres across 138 species and 2,500 natural strain isolates, revealing remarkable centromere variation. Through simulations and in vivo centromere function experiments, we show that a combination of selection, drift, and sexual reproduction plays a crucial role in driving these changes. We also show that the kinetochore interface places constraints on what changes are tolerated.
Our study challenges the traditional view of centromere drive as the sole driver of centromere evolution. We propose a multi-faceted model involving drift and selection during both mitosis and meiosis, shaped by constraints imposed by the kinetochore interface.
This study is a collaboration with the Sherlock lab
Mikus and Dey
Ultrastructure Expansion Microscopy (U-ExM) of the Fission Yeast Schizosaccharomyces pombe
Methods in Molecular Biology, 2025 DOI: 10.1007/978-1-0716-4168-2_4
Ultrastructure expansion microscopy (U-ExM) enables super-resolution imaging of full 3D volumes at increased throughput using conventional microscopes and can be readily combined with commonly used antibodies, dyes, and stains.
In this protocol, we present its application to the fission yeast Schizosaccharomyces pombe.
Mikus F, Ramos A R, Shah H et al.
Charting the landscape of cytoskeletal diversity in microbial eukaryotes
Cell, 2025 DOI: 10.1016/j.cell.2025.09.027
bioRxiv, 2024 DOI: 10.1101/2024.10.18.618984
In this study, we investigate cytoskeletal diversity at an unprecedented scale across the eukaryotic tree of life using Ultrastructure Expansion Microscopy (U-ExM). We carry out high-resolution volumetric imaging of over 200 cultured planktonic eukaryotes across major lineages. By combining U-ExM with pan- and specific immuno-labelling, we reveal novel microtubule and centrin-containing elements and assign molecular identities to enigmatic cytoskeletal structures observed previously only by electron microscopy.
We demonstrate that U-ExM can be used to investigate environmental samples which usually contain mixtures of different eukaryotes, thereby paving the way for cell biology at ultrastructural resolution and unprecedented scale, a crucial step towards understanding and protecting complex ecosystems in the face of biodiversity loss.
This study is a collaboration between our lab and the Dudin lab, Schwab lab, Guichard/Hamel lab, and Husnik lab.
This study would never have been possible without samples from the TREC Mobile Lab Services, Roscoff Culture Collection and Basque microalgae Culture Collection
Ansel M et al.
Origin and evolution of microvilli
Biology of the Cell, 2024 DOI:10.1111/boc.202400054
In this review, we investigate the origins of finger-like, straight, and stable cellular protrusions called microvilli. These structures are found across the metazoan tree of life and mediate processes like nutrient absorption, photosensation, and mechanosensation.
We describe mammalian microvilli in detail, compare them to filopodia, and present a phylogenetic profile of key microvillar proteins. Using these results, we present a hypothesis that microvilli evolved from from filopodia-like ancestral structures in unicellular precursors of animals.
This study is a collaboration with the lab of Thibaut Brunet
Mylan was a visiting student in our lab for five months optimizing U-ExM in Choanoflagellates
We show that the number of kinetochore-microtubule attachment points at centromeres impose a constraint on chromosome number evolution. An insufficient number of attachments result in a force imbalance in the metaphase spindle, declustering kinetochores.
Our work provides an experimental example of how the mechanics of a core cellular process can constrain evolutionary processes.
This preprint is a result from the close collaboration between us and the lab of Gavin Sherlock.
Reza H et al.
Expansion microscopy reveals unique ultrastructural features of pathogenic budding yeast species
Journal of Cell Science, 2024 DOI: 10.1242/jcs.262046
bioRxiv, 2024 DOI: 10.1101/2024.02.20.581313
We establish ultrastructural expansion microscopy (U-ExM) in seven pathogenic yeasts including Candida albicans. Using protein pan-labelling and immunofluorescence of native and tagged proteins, we outline organelle dynamics through the cell cycle. U-ExM uncovers differences in SPB and spindle organisation and dynamics in C. albicans compared to model budding yeast S. cerevisiae.
Hashim was a scientific visitor with the lab for three months learning and optimising U-ExM for a variety of yeasts.
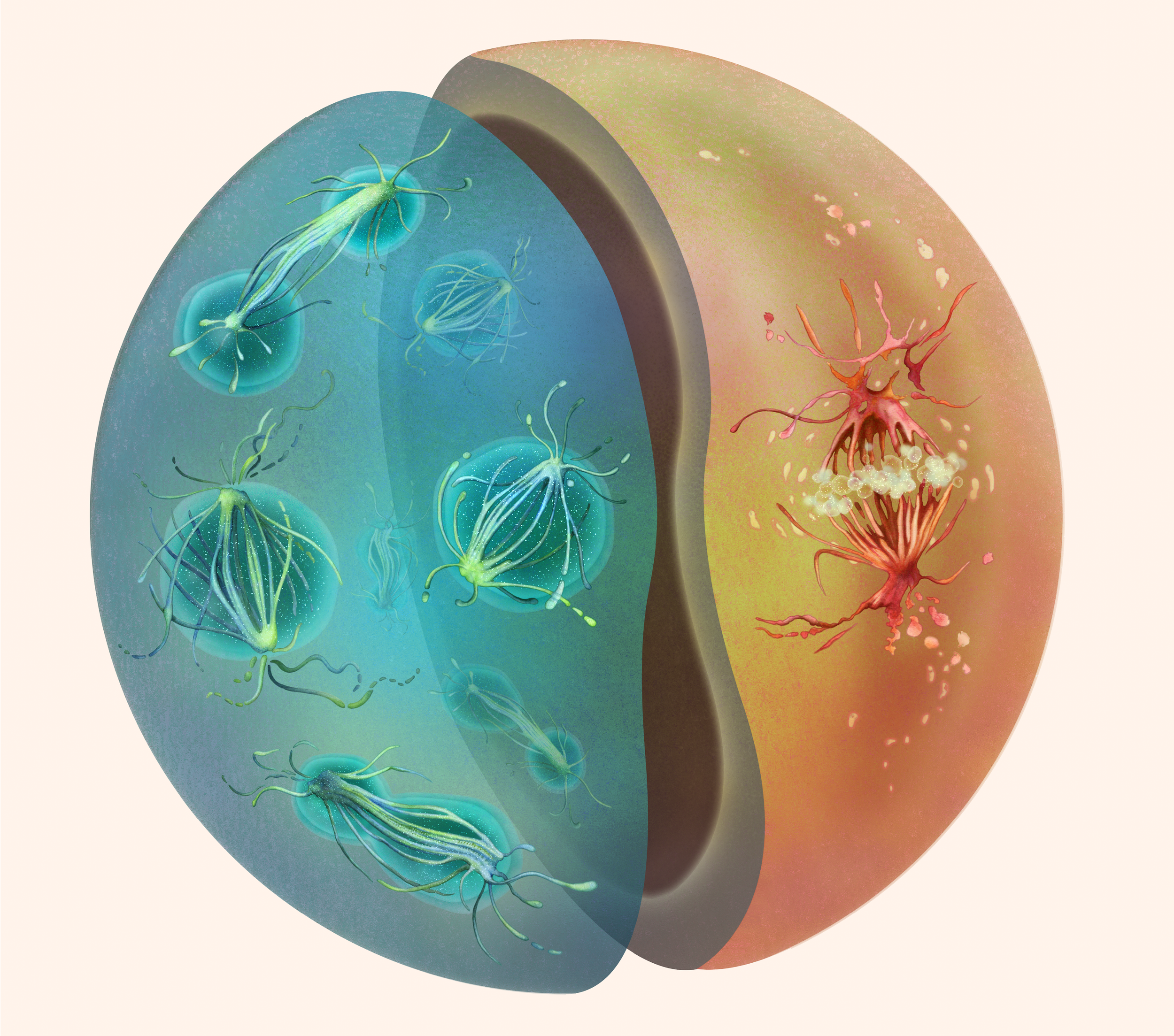
Sphaeroforma arctica and Chromosphaera perkinsii undergoing mitosis, depicted as two halves of a cell, rendered in Haeckel-inspired tones and a naturalist style.
© Nirupa Rao.
Remodelling of the nuclear envelope remodelling during cell division can be vastly different between eukaryotes. Some disassembling their envelope, some divide using an intranuclear spindle, but the evolutionary reasons for adopting one or the other are unclear. Here, we use an integrated comparative genomics and ultrastructural imaging approach to investigate mitotic strategies in Ichthyosporea, close relatives of animals and fungi. Species within this clade have diverged towards either a fungal-like closed or an animal-like open mitosis, most likely to support distinct multi- or uninucleated states. Our results suggest that multinucleated life cycles favour the evolution of closed mitosis.
This preprint emerged from a close collaboration between us and the labs of Omaya Dudin, Yannick Schwab, and Iva Tolić.
Evolutionary cell biology explores the origins, principles, and core functions of cellular features and regulatory networks through the lens of evolution. This emerging field relies heavily on comparative experiments and genomic analyses that focus exclusively on extant diversity and historical events, providing limited opportunities for experimental validation. In this opinion article, we explore the potential for experimental laboratory evolution to augment the evolutionary cell biology toolbox, drawing inspiration from recent studies that combine laboratory evolution with cell biological assays. Primarily focusing on approaches for single cells, we provide a generalizable template for adapting experimental evolution protocols to provide fresh insight into long-standing questions in cell biology.
Here, by using mutant cells exhibiting altered attachment behaviours of mitochondria to microtubules, we show their important role in determining division symmetry. Changes in the attachment of Mts and mitochondria resulted in errors in nuclear positioning and asymmetric cell division. To allow for a quantification of contact sites, we were happy to contribute U-ExM data for all three strains.
Here, we present an ultrastructure expansion microscopy (U-ExM) protocol optimised for use in the model yeasts S. cerevisiae and S. pombe. Increasing the sample size by a factor of roughly four allows for 3D super resolution on conventional microscopy systems. For this publication, we use it to investigate the spatial organisation of the spindle pole body (SPB) and count nuclear pore complexes (NPCs). Combining those specific labels with a general protein staining enables an investigation of subcellular structures within a cellular context.
A major additional improvement was presented by using high pressure freezing instead of chemical fixation methods to preserve structures in a near native state. This further enabled the detection of several proteins with specific antibodies that, in chemical fixations, did not yield sufficient signals - such as for tubulin.
In this review, we discuss the cellular properties and processes that contribute to nuclear size and shape control, drawing examples from across eukaryotes and highlighting conserved themes and pathways. We then outline physiological roles that have been uncovered for specific nuclear morphologies and disease pathologies associated with aberrant nuclear morphology.
Fraser N, Brierley L, Dey G, Polka JK, Pálfy M, Nanni F, Coates JA.
The evolving role of preprints in the dissemination of COVID-19 research and their impact on the science communication landscape.
PLoS Biol. 2021 Apr 2;19(4):e3000959. DOI: 10.1371/journal.pbio.3000959.
Over 125,000 COVID-19-related scientific articles were released within 10 months of the first confirmed case, of which more than 30,000 were hosted by preprint servers. We investigated the attributes of COVID-19 preprints, their access and usage rates, as well as characteristics of their propagation on online platforms. Our data provide evidence for increased scientific and public engagement with preprints related to COVID-19 (COVID-19 preprints are accessed more, cited more, and shared more on various online platforms than non-COVID-19 preprints), as well as changes in the use of preprints by journalists and policymakers. We also find evidence for changes in preprinting and publishing behaviour: COVID-19 preprints are shorter and reviewed faster.
All eukaryotic cells must partition the nuclear envelope in lockstep with cell division at the end of each cell cycle. Cells have evolved a wide range of strategies to carry out nuclear division. We discuss shared and divergent features of these mitotic strategies and speculate on their evolutionary origins.
Before EMBL
Dey G, Culley S, Curran S, et al.
Closed mitosis requires local disassembly of the nuclear envelope.
Nature. 2020 Sep;585(7823):119-123. DOI: 10.1038/s41586-020-2648-3.
We show that ‘closed’ mitosis in the fission yeast Schizosaccharomyces pombe occurs via local disassembly of the nuclear envelope within the narrow bridge connecting segregating daughter nuclei; we identify a key role for Les1 in this process, which restricts nuclear envelope breakdown to the bridge. We find that the regulatory logic of ‘local’ NEB mirrors that of NEB in open mitosis, hinting at the existence of universal NE remodelling mechanisms with implications for our understanding of eukaryotic evolution.
The hyperthermophilic archaeon Sulfolobus is a close relative of the first eukaryotes, and uses ESCRTIII to divide. We constructed the world’s first 75C microscope to image Sulfolobus cells dividing live.
Loss of gene function is common throughout evolution, even though it often leads to reduced fitness. Using experimental evolution of hundreds of budding yeast strains, we found that gene loss can be compensated and can ultimately even facilitate and enhance adaptation.
Newly discovered archaeal genomes encode homologs of key eukaryotic gene families, including cytoskeletal regulators and small GTPases. What does that mean for the cell biology of the archaeal ancestors of eukaryotes, and will these genomes shed light on the evolution of the first eukaryotes, over 2 billion years ago?
Dey G, Jaimovich A, Collins SR, Seki A, Meyer T.
Systematic Discovery of Human Gene Function and Principles of Modular Organization through Phylogenetic Profiling.
Cell Reports. 2015 Feb;10(6):993-1006. DOI: 10.1016/j.celrep.2015.01.025.
Genes that share an evolutionary history are likely to share molecular functions and cellular roles. We make use of this principle to identify co-evolving modules in the human genome, and help assign functions to uncharacterised human genes.
Dey G, Gupta GD, Ramalingam B, et al.
Exploiting cell-to-cell variability to detect cellular perturbations. Plos one. 2014 ;9(3):e90540. DOI: 10.1371/journal.pone.0090540.
Genome-wide functional screens can be incredibly powerful tools, but come with a unique set of challenges, including high levels of variability that can mask subtle results. We develop an algorithm to exploit the variability between cells to detect subtle perturbations in an RNAi screen for endocytosis.