Our Research
We investigate the fundamental organisational principles and evolutionary dynamics of the nuclear compartment across eukaryotes using comparative genomics, cell biology and experimental evolution.
Read about some of our ongoing projects below!
State-of-the-art tool development
In the Dey lab we believe that the fission yeast Schizosaccharomyces pombe is ideally suited to investigate particular aspects of nuclear biology. We are implementing state-of-the-art molecular biology techniques that will allow us to unlock the model’s full potential. We have been busy developing methods for three- and four-color live cell imaging, inducible expression and acute protein degradation to be used in fission yeast.
We have optimised two strategies for 3-colour live imaging:
By using a HALO tag with JF646 ligands we could combine existing strains already tagged with GFP and mCherry with a third marker. Suitable for imaging dynamic processes.
With the mTFP1, mCitrine and E2-Crimson we found good spectral separation. To avoid bleed-through, detection has to be made using a system with gated spectral detection. Suitable to capture precise localization.
We are generally interested in how evolution shapes cell biology. As part of our broader efforts, we are developing screens and reporter tools to be used across evolutionarily close and distant organisms such as S. pombe, the budding yeast Saccharomyces cerevisiae, and the slime mould Physarum polycephalum.
An important component of our toolkit is ultrastructure expansion microscopy (U-ExM). We have already optimised the method for its use in the model yeasts S. pombe and S. cerevisiae (together with the Centriole Lab and Loewith Lab) and are aiming to adjust the protocol to work on complex samples containing multiple species.
Homeostasis in symmetric division
During nuclear division in closed mitosis, nuclear envelope scission must be locally controlled to prevent the leakage of nuclear material. In fission yeast, the separation of nuclei is regulated via the disassembly of a subset of nuclear pore complexes (NPCs) localised to the bridge that connects the two nuclei throughout mitosis. Their gradual removal triggers a localised nuclear envelope breakdown (NEBD) in a process closely resembling NPC remodelling in open mitosis of animal cells (Dey et al., 2020).
How is this disassembly process controlled, and how is it restricted to just a subset of all NPCs? More generally, how is the synthesis and turnover of NPC components regulated through the cell cycle in a system undergoing closed mitosis? The mid zone might furthermore might create a compartment used to remove nuclear proteins and to facilitate bulk exclusion from the nucleoplasm and enable daughter cell rejuvenation.
We are addressing these questions in S. pombe using state-of-the-art light microscopy (together with the EMBL ALMF and the Centriole Lab in Geneva) coupled to acute genetic and chemical perturbations.
Understanding Opisthokont nuclear organization using orthology-based reconstruction of regulatory networks
The nucleus is universally present across eukaryotes but exhibits a great variation in how it is structurally organised. Despite significant advances in genome sequencing, computational tools, and microscopy, little is known about the principles underlying the evolutionary history of nuclear structural organisation that can explain its observed extant diversity. What is known through studies in tractable model systems is difficult to generalise given the limited phylogenetic and phenotypic coverage.
We propose to bridge this gap in knowledge by setting up a workflow to infer protein orthologs using Hidden Markov Models, identify protein interaction modules, and create genotype-to-phenotype maps in uncharacterised Opisthokonts based on data from model systems.
This will also be a valuable resource for evolutionary cell biologists.
This project is a collaboration with Yannick Schwab (EMBL), Omaya Dudin (EPFL), and Tom Williams (University of Bristol)
In the face of different selection pressures, some eukaryotes have evolved to replicate genetic material without generating new daughter cells, resulting in multinucleate cells. The dynamics and synchronicity of nuclear divisions in multinucleate cells are hypothesized to balance replication speed with resource availability, but the factors and mechanisms regulating these divisions are not fully understood.
We want to understand how the environment and lifestyle of an organism shape the nuclear cycle control network. Do changes to the local environment alter the dynamics of nuclear cycles? Which mechanisms control nuclear multiplication in response to environmental cues? Is this regulation conserved between two organisms with different ecological niches?
To address these questions, we will take a comparative approach using two evolutionarily distant unicellular eukaryotes, the free-living slime mold Physarum polycephalum and the malaria parasite Plasmodium falciparum, which both have multinucleated life stages. We will compare the plasticity of nuclear cycling in these organism by using a combination of spatial transcriptomics, high-resolution imaging, basic cell biology techniques, and molecular genetics.
This project is supported by the Health + Life Science Alliance Heidelberg Mannheim awarded to Marie Jacobovitz, and is being carried out in collaboration with Markus Ganter at Heidelberg University Hospital.
Influence of environment on nuclear cycles
Interplay of genomic and nuclear organisation
Eukaryotic genomes show striking differences in karyotype, both in terms of chromosome number and genome size. A well-known example of rapid karyotype evolution is found in muntjacs, where more than 20 chromosome fusion events have reduced the number of chromosomes from 2n = 46 to 2n = 6/7 within species of the same genus. Regardless, each cell cycle, the cell division machinery must rapidly and reproducibly duplicate and partition the cell’s chromosomes, in a process carefully coordinated with growth, nuclear dynamics, and cell cycle signalling.
In this project, we use budding (S. cerevisiae) and fission yeast (S. pombe), two species with similar genome sizes but vastly different chromosome numbers to seek to understand if the cell division machinery is impacted by drastic changes in karyotype. To systematically evaluate the effect of chromosome number, we are using a set of budding yeast strains in which the sixteen native chromosomes have been successively fused.
In parallel, we are using artificial chromosomes with minimal transcriptional potential to evaluate the effect of genome size. By combining experimental evolution (led by the Sherlock Lab at Stanford) and cell biological profiling (led by the Dey Group at EMBL), we are exploring how the cell division machinery is connected to a cell’s karyotype, and how cells are able to adapt to such changes. This project is funded by an EMBL/Stanford Bridging Excellence Fellowship awarded to Jana Helsen.
Coordination of microtubule organizing centers in space and time
Microtubule organizing centers (MTOCs) are key eukaryotic structures for microtubule nucleation and organization and can broadly be categorized into those with and those without a centriole. Centrioles fulfill two critical functions. In mitosis, they localize to the mitotic spindle pole aiding its organization. After cell division, centrioles can be repurposed as basal bodies at the plasma membrane where they are essential for flagella or cilia formation.
The function of centrioles/basal bodies (CBBs) correlates with subcellular localization, but is the spatiotemporal control required for this organelle to fulfill two distinct functions? How did the ancestral CBB evolve these functions? And why are different MTOCs not present simultaneously in a cell?
To address these questions, we will analyze and compare CBB repurposing strategies in two evolutionary distinct eukaryotes, the green alga Chlamydomonas reinhardtii and the slime mold Physarum polycephalum using state-of-the-art microscopy techniques. By interfering with the spatial and temporal coordination of the CBB we will try to elucidate the role of the subcellular location of this bifunctional organelle. Further, we will study how cells cope with several different MTOCs (centriolar and non-centriolar) at a time.
This project is supported by a Marie Sklodowska-Curie postdoctoral fellowship awarded to Caroline Simon and is being carried out in collaboration with the group of Niccolò Banterle at EMBL Heidelberg.
Impact of coenocytic lifestyle on the choice of mitotic strategies
Eukaryotes have evolved a spectrum of mitotic strategies, from open mitosis in animals which break down the nuclear envelope (NE) to closed mitosis in fungi that remodel the nucleus into a dumbbell before partitioning it. With most of the information gathered from a handful of model organisms, the factors driving this incredible diversity in nuclear reorganisation remain largely unknown.
We are interested in whether a coenocytic lifestyle, where many nuclei share a common cytoplasm, favours a more closed mitotic strategy. Does keeping the nuclear envelope intact allow many nuclei to exist and divide simultaneously in the same cell? Is there some interplay between nuclear envelope integrity and the nature of the microtubule organising centre (MTOC) and spindle architecture?
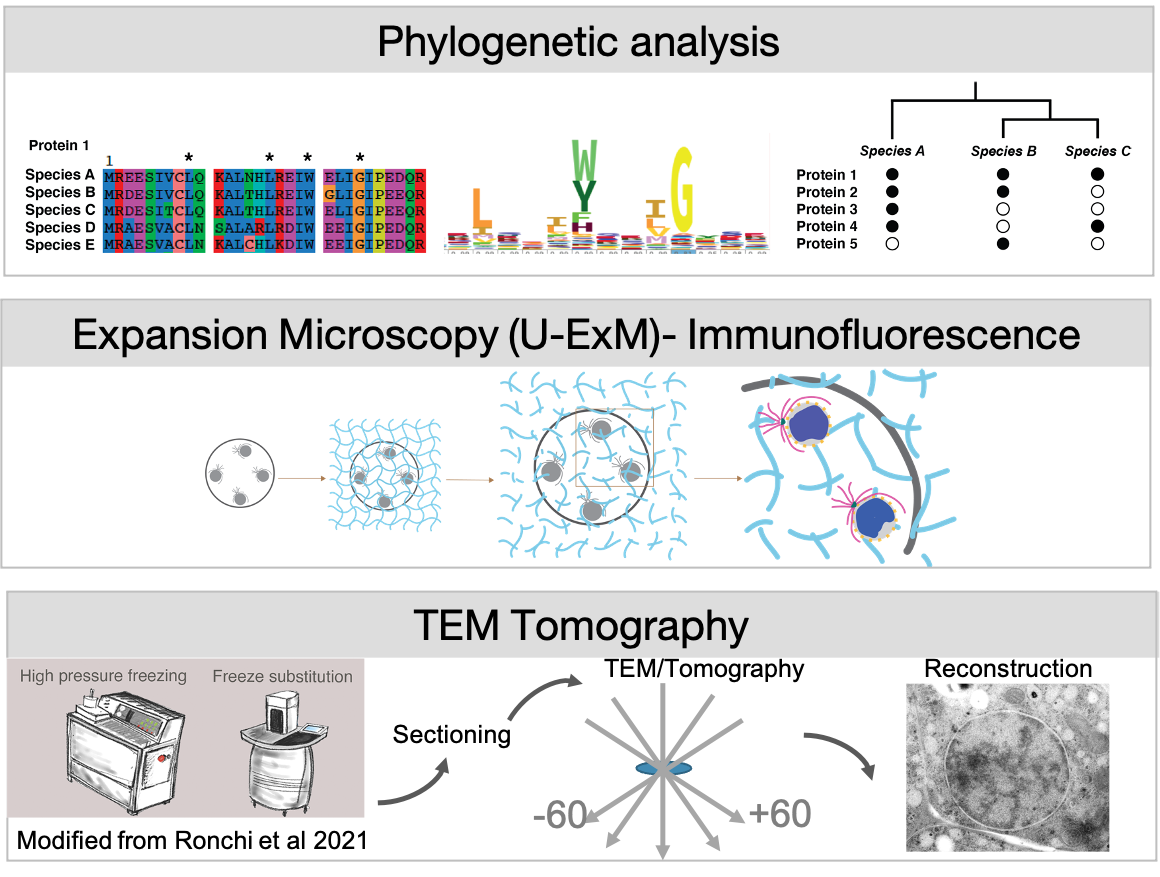
We address these questions in Ichthyosporea, close relatives of animals that exhibit a coenocytic lifestyle. Their unique position on the eukaryotic tree between our favourite animal and fungal models allows us to identify shared mechanisms of nuclear reorganisation. Using phylogenetics, and expansion microscopy and TEM-tomography we compare mitotic strategies between different Ichthyosporean species.
This project is supported by an EIPOD4 postdoctoral fellowship awarded to Hiral Shah, and is being carried out in collaboration with Yannick Schwab at EMBL and Omaya Dudin at EPFL, Lausanne.
TREC and Plan-ExM
We are joining the EMBL TREC (Traversing European Coastlines) expedition in 2023 and 2024!
We aim to create an atlas of planktonic ultrastructural diversity using Expansion Microscopy (ExM). By coupling expansion microscopy (Cryo-ExM and U-ExM) to 18s rRNA FISH we aim to identify species and image them at unprecedented 3D spatial resolution, with a long-term view to assess the repercussions of climate change on free-living microplankton populations.
To achieve this, we are pooling the expertise of several labs within (Schwab team, Saka Group) and outside of EMBL (Dudin Lab, Centriole Lab) .